Discovery of Novel Targets for Cancer Immunotherapy
See manuscripts in Cell (2021), Cell (2018), Current Opinion Immunology (2016), Nature (Feb 2014)
We are using an integrated approach involving genetic screens, scRNA-seq analysis and computational biology approaches to discover novel targets for therapeutic intervention. We frequently use an approach that starts with a human dataset, followed by mechanistic investigation of the identified pathway in murine models. Alternatively, targets discovered in murine models are validated by computational analysis of rich datasets now available from clinical trials of cancer immunotherapies.
See manuscripts in Cell (2021), Cell (2018), Current Opinion Immunology (2016), Nature (Feb 2014)
We are using an integrated approach involving genetic screens, scRNA-seq analysis and computational biology approaches to discover novel targets for therapeutic intervention. We frequently use an approach that starts with a human dataset, followed by mechanistic investigation of the identified pathway in murine models. Alternatively, targets discovered in murine models are validated by computational analysis of rich datasets now available from clinical trials of cancer immunotherapies.
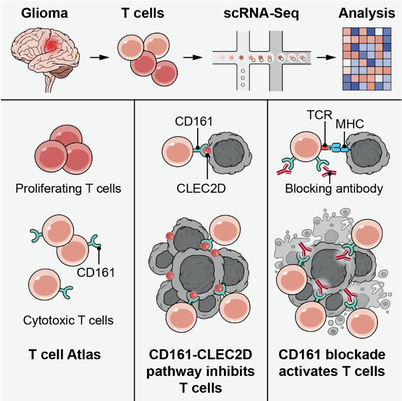
Discovery of CD161 as a target for cancer immunotherapy.
For example, we have performed an in depth analysis of T cells in human diffuse gliomas which led to the discovery of an interesting inhibitory receptor for tumor-infiltrating T cells. T cells are critical effectors of cancer immunotherapies, but little is known about their gene expression programs in human diffuse gliomas. We leveraged single-cell RNA-seq to chart the gene expression and clonal landscape of tumor-infiltrating T cells across 31 patients with IDH-wildtype glioblastoma and IDH-mutant glioma. We identified potential effectors of anti-tumor immunity in subsets of T cells that co-expressed cytotoxic programs and several NK cell genes. Analysis of clonally expanded tumor-infiltrating T cells further identified the NK gene KLRB1 (encoding CD161) as a candidate inhibitory receptor. Accordingly, genetic inactivation of KLRB1 or antibody-mediated CD161 blockade enhanced T cell-mediated killing of glioma cells in vitro and their anti-tumor function in vivo. KLRB1 and its associated transcriptional program are also expressed by substantial T cell populations in other human cancers. Our work provides an atlas of T cells in gliomas and highlights CD161 and other NK cell receptors as immunotherapy targets.
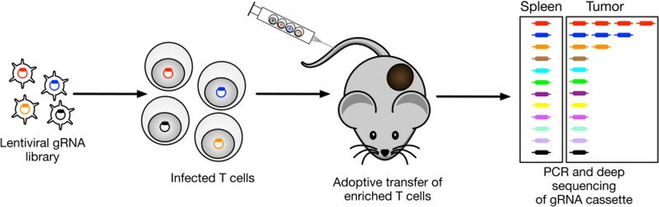
We also use systematic genetic screens to discover key regulators of anti-tumor immunity. For example, we developed a novel approach for in vivo discovery of genes that inhibit T cell function in the tumor microenvironment. In a normal immune response, T cells proliferate extensively following antigen recognition, but this physiological response is severely blunted by multiple immunosuppressive mechanisms in the tumor microenvironment. We reasoned that gRNAs targeting critical negative regulators in T cells can restore their proliferative response to tumor antigen recognition. We transduced CD8 T cells with pooled gRNA libraries and transferred these T cells into tumor-bearing mice. Seven to ten days later, T cells were isolated to quantify the representation of gRNAs in tumors and control tissues by deep sequencing. We have discovered a large number of genes that inhibit T cell function in the tumor microenvironment and are now investigating the molecular mechanisms by which these genes constrain T cell function in tumors.
We also use systematic genetic screens to discover key regulators of anti-tumor immunity. For example, we developed a novel approach for in vivo discovery of genes that inhibit T cell function in the tumor microenvironment. In a normal immune response, T cells proliferate extensively following antigen recognition, but this physiological response is severely blunted by multiple immunosuppressive mechanisms in the tumor microenvironment. We reasoned that gRNAs targeting critical negative regulators in T cells can restore their proliferative response to tumor antigen recognition. We transduced CD8 T cells with pooled gRNA libraries and transferred these T cells into tumor-bearing mice. Seven to ten days later, T cells were isolated to quantify the representation of gRNAs in tumors and control tissues by deep sequencing. We have discovered a large number of genes that inhibit T cell function in the tumor microenvironment and are now investigating the molecular mechanisms by which these genes constrain T cell function in tumors.
Design of in vivo CRISPR screen for discovery of negative regulators of T cell function in the tumor microenvironment.
Mathewson, N. D., Ashenberg, O., Tirosh, I., Gritsch, S., Perez, E. M., Marx, S., Jerby-Arnon, L., Chanoch-Myers, R., Hara, T., Richman, A. R., Ito, Y., Pyrdol, J., Friedrich, M., Schumann, K., Poitras, M. J., Gokhale, P. C., Gonzalez Castro, L. N., Shore, M. E., Hebert, C. M., Shaw, B., Cahill, H. L., Drummond, M., Zhang, W., Olawoyin, O., Wakimoto, H., Rozenblatt-Rosen, O., Brastianos, P. K., Liu, X. S., Jones, P.S., Cahill. D. P., Frosch, M. P., Louis, D.N., Freeman, G. J., Ligon, K. L., Marson, A., Chiocca, E. A., Reardon, D. A., Regev, A., Suvà, M. L., Wucherpfennig, K. W. (2021) Inhibitory CD161 Receptor Identified in Glioma-infiltrating T cells by Single Cell Analysis. Cell, online Feb. 15 2021
Jerby-Arnon, L., Shah, P., Cuoco, M. S., Rodman, C., Su, M. J., Melms, J. C., Leeson, R., Kanodia, A., Mei, S., Lin, J. R., Wang, S., Rabasha, B., Liu, D., Zhang, G., Margolais, C., Ashenberg, O., Ott, P. A., Buchbinder, E. I., Haq, R., Hodi, F. S., Boland, G. M., Sullivan, R. J., Frederick, D. T., Miao, B., Moll, T., Flaherty, K. T., Herlyn, M., Jenkins, R. W., Thummalapalli, R., Kowalczyk, M. S., Canadas, I., Schilling, B., Cartwright, A. N. R., Luoma, A. M., Malu, S., Hwu, P., Bernatchez, C., Forget, M. A., Barbie, D. A., Shalek, A. K., Tirosh, I., Sorger, P. K., Wucherpfennig, K., Van Allen, E. M., Schadendorf, D., Johnson, B. E., Rotem, A., Rozenblatt Rosen, O., Garraway, L. A., Yoon, C. H., Izar, B., and Regev, A. (2018) A Cancer Cell Program Promotes T Cell Exclusion and Resistance to Checkpoint Blockade. Cell 175, 984-997 e924
Wucherpfennig, K. W., and Cartwright, A. N. (2016) Genetic screens to study the immune system in cancer. Curr Opin Immunol 41, 55-61
Zhou, P., Shaffer, D. R., Alvarez Arias, D. A., Nakazaki, Y., Pos, W., Torres, A. J., Cremasco V., Dougan, S. K., Cowley, G. S., Elpek, K., Brogdon, J., Lamb, J., Turley, S. J., Ploegh, H. L., Root, D. E., Love, J. C., Dranoff, G., Hacohen, N., Cantor, H., and Wucherpfennig, K. W. (2014) In vivo discovery of immunotherapy targets in the tumour microenvironment. Nature 506, 52-57
Jerby-Arnon, L., Shah, P., Cuoco, M. S., Rodman, C., Su, M. J., Melms, J. C., Leeson, R., Kanodia, A., Mei, S., Lin, J. R., Wang, S., Rabasha, B., Liu, D., Zhang, G., Margolais, C., Ashenberg, O., Ott, P. A., Buchbinder, E. I., Haq, R., Hodi, F. S., Boland, G. M., Sullivan, R. J., Frederick, D. T., Miao, B., Moll, T., Flaherty, K. T., Herlyn, M., Jenkins, R. W., Thummalapalli, R., Kowalczyk, M. S., Canadas, I., Schilling, B., Cartwright, A. N. R., Luoma, A. M., Malu, S., Hwu, P., Bernatchez, C., Forget, M. A., Barbie, D. A., Shalek, A. K., Tirosh, I., Sorger, P. K., Wucherpfennig, K., Van Allen, E. M., Schadendorf, D., Johnson, B. E., Rotem, A., Rozenblatt Rosen, O., Garraway, L. A., Yoon, C. H., Izar, B., and Regev, A. (2018) A Cancer Cell Program Promotes T Cell Exclusion and Resistance to Checkpoint Blockade. Cell 175, 984-997 e924
Wucherpfennig, K. W., and Cartwright, A. N. (2016) Genetic screens to study the immune system in cancer. Curr Opin Immunol 41, 55-61
Zhou, P., Shaffer, D. R., Alvarez Arias, D. A., Nakazaki, Y., Pos, W., Torres, A. J., Cremasco V., Dougan, S. K., Cowley, G. S., Elpek, K., Brogdon, J., Lamb, J., Turley, S. J., Ploegh, H. L., Root, D. E., Love, J. C., Dranoff, G., Hacohen, N., Cantor, H., and Wucherpfennig, K. W. (2014) In vivo discovery of immunotherapy targets in the tumour microenvironment. Nature 506, 52-57